Kruskal-Wallis is (almost) a one-way ANOVA on ranked data
Jonas Kristoffer Lindeløv
This document presents the close relationship between the Kruskal-Wallis test and one-way ANOVAs. Namely, that Kruskal-Wallis test is, to a close approximation, just a one-way ANOVA on \(rank\)ed \(y\). It is an appendix to the post “Common statistical tests as linear models”.
TL;DR: Below, I argue that this approximation is good enough when the sample size is 12 or greater and virtually perfect when the sample size is 30 or greater.
1 Simulate data
Calculate statistics for different sample sizes (\(N\)) and differences between groups (\(mu\)).
library(tidyverse)
# Parameters
Ns = c(seq(from=6, to=20, by=2), 30, 50, 80)
mus = c(0, 0.5, 1) # Means
PERMUTATIONS = 1:200
# Run it
D = expand.grid(set=PERMUTATIONS, mu=mus, N=Ns) %>%
mutate(
# Generate data. One normal and one weird
value = map2(N, mu, ~c(rnorm(.x), rnorm(.x) + .y, rnorm(.x))),
group = map(N, ~factor(c(rep('A', .x), rep('B', .x), rep('C', .x)))),
# Tests
kruskal_raw = map2(value, group, ~ kruskal.test(.x ~ .y)),
anova_raw = map2(value, group, ~aov(rank(.x) ~ .y)),
# Tidy it up
kruskal = map(kruskal_raw, broom::tidy),
anova = map(anova_raw, broom::glance)
) %>%
# Get as columns instead of lists; then remove "old" columns
unnest(kruskal, anova, .sep='_') %>%
select(-value, -group, -kruskal_raw, -anova_raw)
head(D)2 Show results
I have found that the effect size (\(mu\)) makes no difference, so I collapse the analyses below across \(mu\).
D$N = factor(D$N) # Make N a factor for prettier plotting
library(ggplot2)
library(patchwork)
# A straight-up comparison of the p-values
p_relative = ggplot(D, aes(x=kruskal_p.value, y=anova_p.value, color=N)) +
geom_line() +
geom_vline(xintercept=0.05, lty=2) +
geom_hline(yintercept=0.05, lty=2) +
labs(title='Absolute relation', x = 'Kruskal-Wallis p-value', y = 'ANOVA p-value') +
#coord_cartesian(xlim=c(0, 0.10), ylim=c(0, 0.11)) +
theme_gray(13) +
guides(color=FALSE)
# Looking at the difference (error) between p-values
p_error_all = ggplot(D, aes(x=kruskal_p.value, y=anova_p.value-kruskal_p.value, color=N)) +
geom_line() +
geom_vline(xintercept=0.05, lty=2) +
labs(title='Error', x = 'Kruskal-Wallis p-value', y = 'T-test p-value deviation') +
theme_gray(13) +
guides(color=FALSE)
# Same, but zoomed in around p=0.05
p_error_zoom = ggplot(D, aes(x=kruskal_p.value, y=anova_p.value-kruskal_p.value, color=N)) +
geom_line() +
geom_vline(xintercept=0.05, lty=2) +
labs(title='Zoomed error', x = 'Kruskal-Wallis p-value', y = 'T-test p-value deviation') +
coord_cartesian(xlim=c(0, 0.10), ylim=c(-0.020, 0.000)) +
theme_gray(13)
# Show it. Patchwork is your friend!
p_relative + p_error_all + p_error_zoom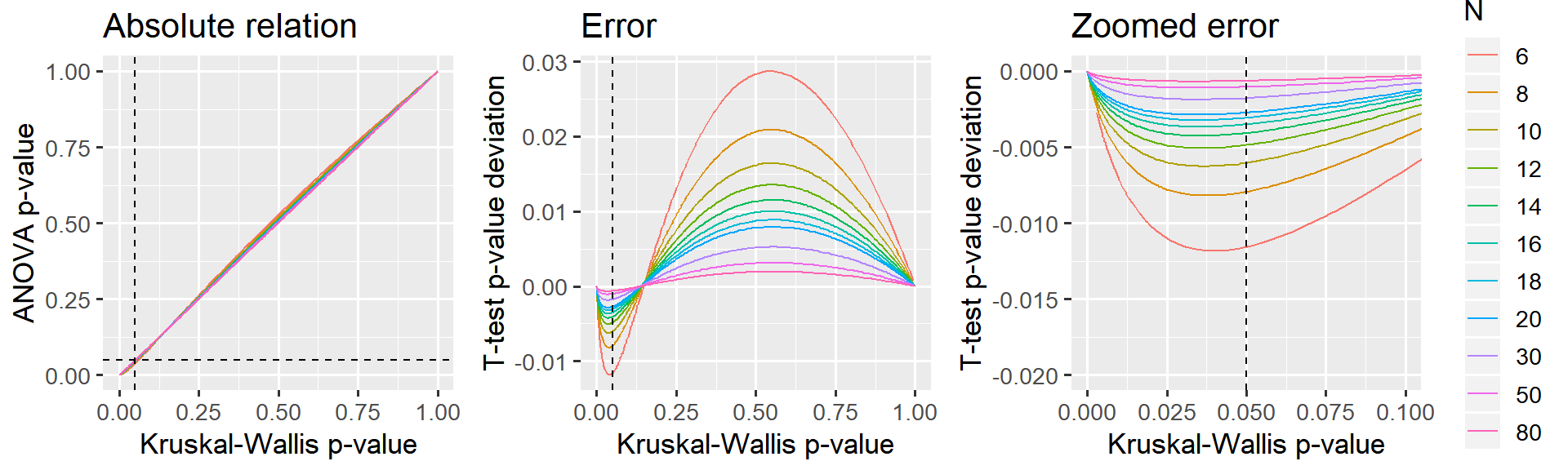
3 Conclusion
Reasonably accurate when N = 12 or greater. Almost exact when N = 30.